PREVALENCE OF PSEUDOMONAS AERUGINOSA IN WOUND INFECTIONS IN KHYBER PAKHTUNKHWA, PAKISTAN
DOI:
https://doi.org/10.71000/q8dx5w91Keywords:
Khyber Pakhtunkhwa, Nosocomial infections, Biochemical tests, Infection control, Risk factors, Pseudomonas aeruginosa, Wound InfectionAbstract
Background: Pseudomonas aeruginosa is a Gram-negative, rod-shaped bacterium and a major cause of hospital-acquired infections globally. Its metabolic adaptability and multidrug resistance make it especially dangerous in immunocompromised individuals. Commonly found in water, soil, and hospital environments, it can cause severe infections including pneumonia, bloodstream infections, and chronic wound infections. Its persistence and resistance have led the World Health Organization to classify it among critical priority pathogens for the development of new antibiotics.
Objective: To determine the prevalence of P. aeruginosa in wound infections across three major teaching hospitals in Khyber Pakhtunkhwa (KPK), Pakistan, and evaluate key epidemiological factors contributing to its occurrence.
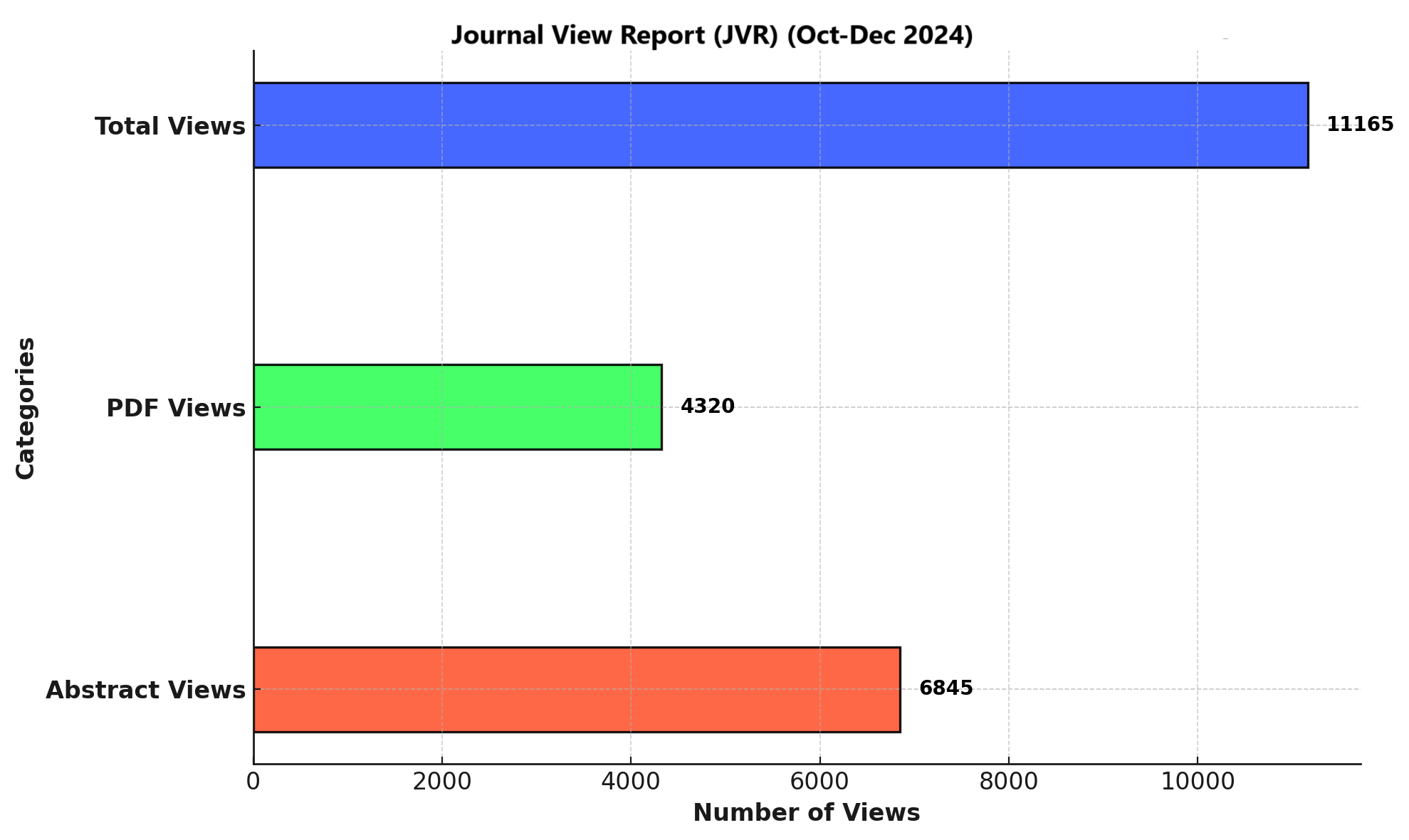
Methods: A cross-sectional study was conducted using 57 wound swab samples collected from Mardan Medical Complex (MMC), Khyber Teaching Hospital (KTH), and Saidu Teaching Hospital (STH). Samples were processed using standard microbiological protocols, including culturing on Blood Agar and MacConkey Agar, followed by Gram staining and biochemical testing (catalase, oxidase, citrate, and indole tests). The data were analyzed with SPSS version 26.0 and Microsoft Excel, with findings expressed in frequencies and percentages.
Results: Out of 57 samples, 22 (38.6%) tested positive for P. aeruginosa. Hospital-wise distribution showed 9 cases (40.9%) at MMC, 7 (31.8%) at KTH, and 6 (27.3%) at STH. All isolates displayed beta-hemolysis on Blood Agar, non-lactose fermentation on MacConkey Agar, and were confirmed as Gram-negative rods. Biochemical testing revealed 100% positivity for catalase and citrate utilization, 95% for oxidase, and 0% for indole. Infections were more frequent in males (63.6%) and in the 41–60 age group (36.4%).
Conclusion: The significant prevalence of P. aeruginosa in wound infections across hospitals in KPK calls for stronger infection control protocols and localized antibiotic stewardship. The higher incidence among males and middle-aged adults highlights potential occupational or comorbid risk factors. Broader regional studies are necessary to assess resistance patterns and support effective clinical decision-making.
References
Lalucat J, Mulet M, Gomila M, García-Valdés E. Genomics in bacterial taxonomy: impact on the genus Pseudomonas. Genes. 2020 Jan 29;11(2):139.
Crone S, Vives‐Flórez M, Kvich L, Saunders AM, Malone M, Nicolaisen MH, Martínez‐García E, Rojas‐Acosta C, Catalina Gomez‐Puerto M, Calum H, Whiteley M. The environmental occurrence of Pseudomonas aeruginosa. Apmis. 2020 Mar;128(3):220-31.
Wei, L., Wu, Q., Zhang, J., Guo, W., Gu, Q., Wu, H., ... & Zeng, X. (2020). Prevalence, virulence, antimicrobial resistance, and molecular characterization of Pseudomonas aeruginosa isolates from drinking water in China. Frontiers in Microbiology, 11, 544653.
Sathe, N., Beech, P., Croft, L., Suphioglu, C., Kapat, A., & Athan, E. (2023). Pseudomonas aeruginosa: Infections and novel approaches to treatment “Knowing the enemy” the threat of Pseudomonas aeruginosa and exploring novel approaches to treatment. Infectious medicine, 2(3), 178-194.
Nasser, M., Gayen, S., & Kharat, A. S. (2020). Prevalence of β-lactamase and antibiotic-resistant Pseudomonas aeruginosa in the Arab region. Journal of global antimicrobial resistance, 22, 152-160.
Jafar, S., Ilahi, N. E., Ullah, A., & Samad, F. U. (2023). 61. Prevalence and antimicrobial susceptibility patterns of extended spectrum β-lactamase producers and non-producers Pseudomonas aeruginosa in Peshawar. Pure and Applied Biology (PAB), 12(1), 769-778.
Irum, S., Naz, K., Ullah, N., Mustafa, Z., Ali, A., Arslan, M., ... & Andleeb, S. (2021). Antimicrobial resistance and genomic characterization of six new sequence types in multidrug-resistant Pseudomonas aeruginosa clinical isolates from Pakistan. Antibiotics, 10(11), 1386.
Diorio-Toth, L., Irum, S., Potter, R. F., Wallace, M. A., Arslan, M., Munir, T., ... & Dantas, G. (2022). Genomic surveillance of clinical Pseudomonas aeruginosa isolates reveals an additive effect of carbapenemase production on carbapenem resistance. Microbiology spectrum, 10(3), e00766-22.
Ali, H., Rizwan, M., Ahmad, S., Khan, Z. U., Fahad, S., & Kashif, M. (2025). Antibiotic Resistance Patterns of Pseudomonas aeruginosa Isolated from Clinical Samples in Khyber Pakhtunkhwa, Pakistan. Journal of Health, Wellness and Community Research, 46-46.
Jafar S, Ilahi NE, Ullah A, Samad FU. 61. Prevalence and antimicrobial susceptibility patterns of extended spectrum β-lactamase producers and non-producers Pseudomonas aeruginosa in Peshawar. Pure and Applied Biology (PAB). 2023 Mar 28;12(1):769-78.
Arooj, I., Asghar, A., Javed, M., Elahi, A., & Javaid, A. (2022). Prevalence and antibiotic susceptibility profiling of MDR pseudomonas aeruginosa from UTI patients of Southern Punjab, Pakistan. RADS Journal of Biological Research & Applied Sciences, 13(1), 1-9.
Al-Bayati, S. S., Al-Ahmer, S. D., Shami, A. M. M., & Al-Azawi, A. H. (2021). Isolation and identification of Pseudomonas aeruginosa from clinical samples. Biochem Cell Arch, 21(2), 3931-3935.
Badri, M. A., Alkhaili, M. A., Aldhaheri, H., Yang, G., Albahar, M., Alrashdi, A., ... & Alhyas, L. (2021). Experiencing the unprecedented COVID-19 lockdown: Abu Dhabi older adults’ challenges and concerns. International Journal of Environmental Research and Public Health, 18(24), 13427.
Li, W., Yang, Z., Hu, J., Wang, B., Rong, H., Li, Z., ... & Xu, H. (2022). Evaluation of culturable ‘last-resort’antibiotic resistant pathogens in hospital wastewater and implications on the risks of nosocomial antimicrobial resistance prevalence. Journal of Hazardous Materials, 438, 129477.
Rukyaa J, Temba L, Kachira P, Mwanansao C, Seni J. Prevalence and antimicrobial susceptibility patterns of bacteria colonizing the external ocular surfaces of patients undergoing ocular surgeries at Bugando Medical Center in Mwanza, Tanzania. BMC Res Notes. 2024;17(1):193.
Ali S, Assafi M. Prevalence and antibiogram of Pseudomonas aeruginosa and Staphylococcus aureus clinical isolates from burns and wounds in Duhok City, Iraq. J Infect Dev Ctries. 2024;18(1):82-92.
Tahmasebi H, Dehbashi S, Nasaj M, Arabestani MR. Molecular epidemiology and collaboration of siderophore-based iron acquisition with surface adhesion in hypervirulent Pseudomonas aeruginosa isolates from wound infections. Sci Rep. 2022;12(1):7791.
Khalili Y, Memar MY, Farajnia S, Adibkia K, Kafil HS, Ghotaslou R. Molecular epidemiology and carbapenem resistance of Pseudomonas aeruginosa isolated from patients with burns. J Wound Care. 2021;30(2):135-41.
Arfaoui A, Rojo-Bezares B, Fethi M, López M, Toledano P, Sayem N, et al. Molecular characterization of Pseudomonas aeruginosa from diabetic foot infections in Tunisia. J Med Microbiol. 2024;73(7).
Alhubail A, Sewify M, Messenger G, Masoetsa R, Hussain I, Nair S, et al. Microbiological profile of diabetic foot ulcers in Kuwait. PLoS One. 2020;15(12):e0244306.
Hoque MN, Jahan MI, Hossain MA, Sultana M. Genomic diversity and molecular epidemiology of a multidrug-resistant Pseudomonas aeruginosa DMC30b isolated from a hospitalized burn patient in Bangladesh. J Glob Antimicrob Resist. 2022;31:110-8.
Azam S, Mustafa MZ, Taj MK, Mengal MA, Mengal MA, Hussain A. Drug susceptibility, Prevalence and effect of Pseudomonas aeruginosa on wound healing of burn patients. J Pak Med Assoc. 2024;74(1):67-71.
Cleland H, Stewardson A, Padiglione A, Tracy L. Bloodstream infections and multidrug resistant bacteria acquisition among burns patients in Australia and New Zealand: A registry-based study. Burns. 2024;50(6):1544-54.
Downloads
Published
Issue
Section
License
Copyright (c) 2025 Hamad Ali, Jahangir Khan, Hammad Khan, Awais Javed, Muhammad Usama, Qaisar Khan, Rikza Mobeen (Author)

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.