PREVALENCE OF ANTIBIOTIC-DEGRADING ENZYME GENES IN HOSPITAL WASTEWATER-ASSOCIATED BACTERIAL COMMUNITIES: A CROSS-SECTIONAL STUDY
DOI:
https://doi.org/10.71000/khw5md07Keywords:
Antibiotic Resistance; Beta-Lactamases; Bacterial Genes; Carbapenem Resistance; Environmental Monitoring; Hospital Wastewater; Polymerase Chain ReactionAbstract
Background: Hospital wastewater is a significant environmental reservoir for antimicrobial resistance, facilitating the dissemination of antibiotic-degrading enzyme genes that pose threats to public health. Understanding the distribution of these genes in effluent streams is essential for surveillance and control strategies.
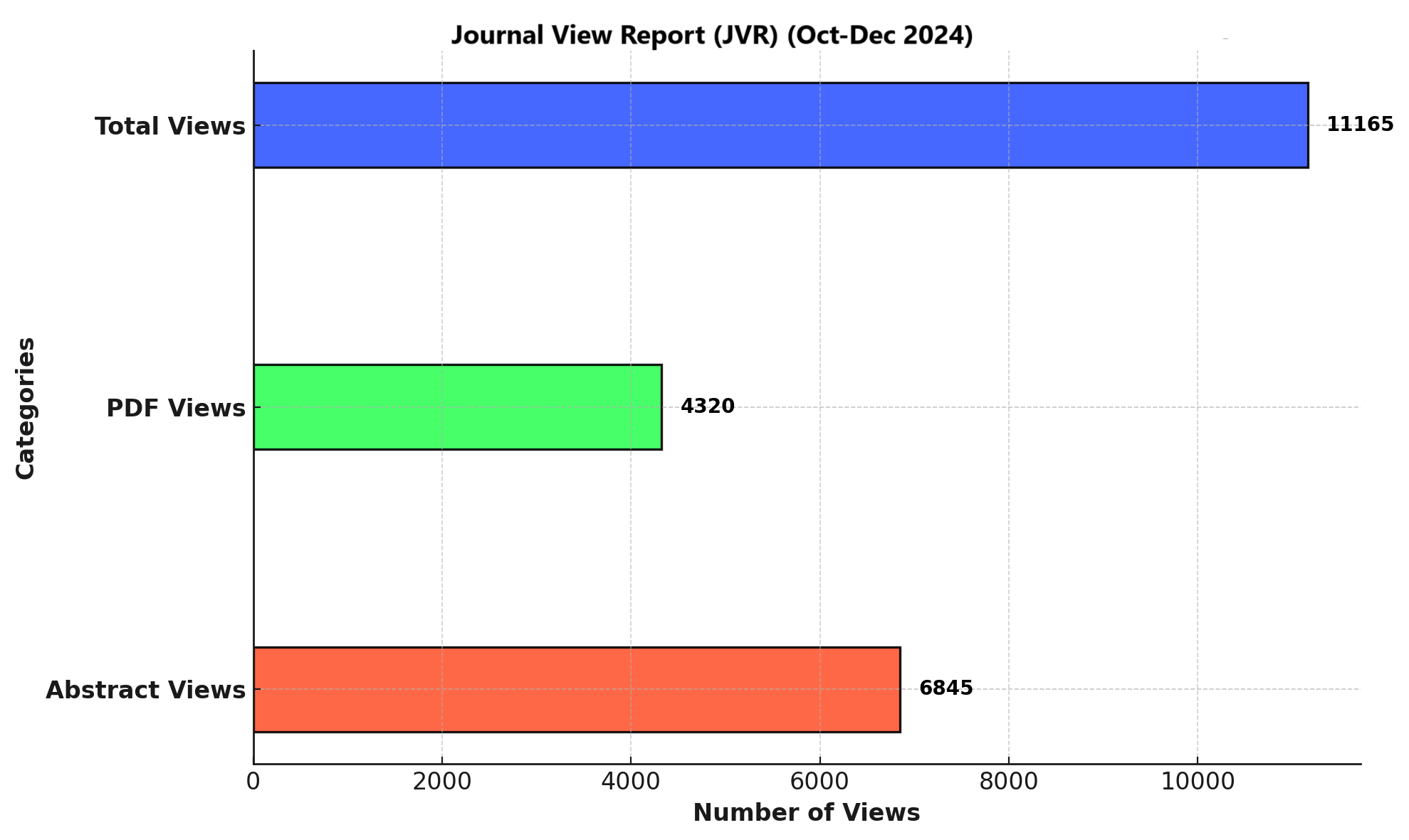
Objective: To determine the prevalence of selected antibiotic-degrading enzyme genes in bacterial isolates recovered from hospital wastewater across multiple collection points.
Methods: A cross-sectional study was conducted over three months in which wastewater samples were collected from five hospital discharge sites. Samples were cultured using standard microbiological techniques, followed by genomic DNA extraction. Polymerase Chain Reaction (PCR) assays were performed to detect key antibiotic-degrading enzyme genes, including blaTEM, blaCTX-M, blaNDM, and aac(6’)-Ib. Data were analyzed to determine gene prevalence and distribution across sampling sites.
Results: Among the cultured bacterial isolates, blaTEM was the most prevalent gene, followed by moderate detection of blaCTX-M and aac(6’)-Ib. A lower frequency of blaNDM was observed, though its presence remains clinically significant due to its association with carbapenem resistance. Gene distribution patterns showed no statistically significant variation among sampling sites, indicating uniform dissemination throughout the wastewater system.
Conclusion: The study highlights the presence of clinically important antibiotic-degrading enzyme genes in hospital wastewater, emphasizing the role of effluent environments in maintaining and spreading antimicrobial resistance. Strengthening wastewater treatment, monitoring programs, and resistance surveillance is critical to minimize environmental dissemination of these genes.
References
Patra M, Dubey SKJRoEC, Toxicology. Antimicrobial resistance transmission in environmental matrices: current prospects and future directions. 2025;263(1):20.
Motlhalamme T, Paul L, Singh V. Environmental Reservoirs, Genomic Epidemiology, and Mobile Genetic Elements. Antimicrobial Resistance: Factors to Findings: Omics and Systems Biology Approaches: Springer; 2024. p. 239-73.
Nass NM, Zaher KAJA. Beyond the Resistome: Molecular Insights, Emerging Therapies, and Environmental Drivers of Antibiotic Resistance. 2025;14(10):995.
Mohanty D, Kumari P, Bera AK, Sahoo AK, Baisakhi B, Kumari M, et al. Emerging Challenges of Extended-Spectrum β-Lactamase Producing Pathogen: Laboratory Strategies for Detection. Laboratory Techniques for Fish Disease Diagnosis: Springer; 2025. p. 555-72.
Zhang H, Luo Y, Zhu X, Ju FJB. Environmental antimicrobial resistance: key reservoirs, surveillance and mitigation under One Health. 2025;1(1).
Barathan M, Ng S-L, Lokanathan Y, Ng MH, Law JXJIJoMS. Unseen weapons: bacterial extracellular vesicles and the spread of antibiotic resistance in aquatic environments. 2024;25(6):3080.
Zambrano MMJARoG. Interplay between antimicrobial resistance and global environmental change. 2023;57(1):275-96.
Fadlalla IM. Perspective Chapter: One Health Approach Impact in Salmonella Public Health–Nexus with Molecular Mechanisms of Antimicrobial Resistance. 2025.
Srivastava S, Singh RJW, Air,, Pollution S. Antibiotic-Resistant Bacterial Diversity in Pharmaceutical Discharge with their Multi-Drug Resistance Capabilities. 2025;236(15):997.
Divya V, Vishwajit B, Adhoni SA, Manawadi S, Rajeshwar V, Khalid M, et al. Impacts of pharmaceutical and personal care products contamination as emerging contaminants in urban ecosystem: emerging risks and future challenges. 2025;37(1):1-18.
Goh JXH, Tan LT-H, Law JW-F, Khaw K-Y, Ab Mutalib N-S, He Y-W, et al. Insights into carbapenem resistance in Vibrio species: current status and future perspectives. 2022;23(20):12486.
Olanrewaju OS, Bezuidenhout CCJFiM. Harnessing beneficial bacteria to remediate antibiotic-polluted agricultural soils: integrating source diversity, bioavailability modulators, and ecological impacts. 2025;16:1635233.
Akunne OZ, Emmanuel BN, Saidu UF, Akinnawo AS, Adeyemi NA, Egoh LC, et al. Antibiotic Resistance: The Growing Threats and Potential Solutions. 2025;4.
Huijbers PM, Bobis Camacho J, Hutinel M, Larsson DJ, Flach C-FJIJoER, Health P. Sampling considerations for wastewater surveillance of antibiotic resistance in fecal bacteria. 2023;20(5):4555.
Ador MAA, Haque MS, Paul SI, Chakma J, Ehsan R, Rahman AJAS. Potential application of PCR based molecular methods in fish pathogen identification: a review. 2021;22(1).
Puljko A. Wastewater as a source for the spread of resistance to third-generation cephalosporins and carbapenems to the aquatic environment: University of Zagreb. Faculty of Food Technology and Biotechnology …; 2024.
Adbaru MG. Characterization of microbial resistome in bacteria isolated from human, environmental and animal sources using DNA microarray technology and genome sequencing: lmu; 2024.
Too RJ. Characterization of Carbapenem-resistant Enterobacterales in Nairobi River, Wastewater Treatment Plant, and Slaughterhouse Discharge in Nairobi County, Kenya: University of Nairobi; 2024.
Aleem M, Azeem AR, Rahmatullah S, Vohra S, Nasir S, Andleeb S, et al. Prevalence of bacteria and antimicrobial resistance genes in hospital water and surfaces. 2021;13(10).
Waśko I, Kozińska A, Kotlarska E, Baraniak AJIjoer, health p. Clinically relevant β-lactam resistance genes in wastewater treatment plants. 2022;19(21):13829.
Adiba RA, Ferdous RM. Characterization of pseudomonas aeruginosa from hospital sewage water and nearby community water based on their multi-drug-resistant gene: Brac University; 2023.
Rahman A. Molecular profiling, antibiotic susceptibility patterns and pathogenic traits of ESBL producing Acinetobacter baumannii Isolated from wastewater discharges from Goranchatbari sub-catchment area in Dhaka city: Brac University; 2024.
Santosaningsih D, Fadriyana AP, David NI, Ratridewi IJTM, Disease I. Prevalence and abundance of beta-lactam resistance genes in hospital wastewater and enterobacterales wastewater isolates. 2023;8(4):193.
Mutuku CS. Occurrence of antimicrobial pharmaceuticals and characterization of β-lactamases in Gram-negative pathogens from wastewater: University of Pécs (Hungary); 2023.
Christopher SM. Occurrence of antimicrobial pharmaceuticals and characterization of β-lactamases in Gram-negative pathogens from wastewater. 2023.
Downloads
Published
Issue
Section
License
Copyright (c) 2025 Zarmeena Gul, Akramullah, Muhammad Amir, Haroon Riaz, Sajjad Ahmad (Author)

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.